Featured products
-

PMSF BioChemica
A0999 -

IPTG BioChemica
A1008 -

DTT BioChemica
A1101 -

NBT BioChemica
A1243 -

-

107 products available
- 2-Nitrophenyl-ß-D-Galactopyranoside BioChemica A1272,0025 · 369-07-3
- 3,3',5,5'-Tetramethylbenzidine BioChemicaA3840,0025 · 54827-17-7
- ABTS® BioChemica A1088,0010 · 30931-67-0
- ABTS® BioChemica A1088,0005 · 30931-67-0
- Acrylamide - Solution (30 %) - Mix 29 : 1 for molecular biologyA4983,0500 ·
- Acrylamide - Solution (30 %) - Mix 37.5 : 1 for molecular biologyA3626,0500 ·
- Acrylamide - Solution (30 %) - Mix 37.5 : 1 for molecular biologyA3626,1000 ·
- Acrylamide - Solution (40 %) - Mix 19 : 1 for molecular biologyA3658,1000 ·
- Acrylamide - Solution (40 %) - Mix 29 : 1 for molecular biologyA0385,1000 ·
- Acrylamide - Solution (40 %) - Mix 29 : 1 for molecular biologyA0385,0500 ·
- Acrylamide - Solution (40 %) - Mix 37.5 : 1 for molecular biologyA4989,0500 ·
- Acrylamide 2K - Solution (18 %) for denaturing DNA-PAGEA7582,9010 ·
- Acrylamide 2K - Solution (8 %) for denaturing DNA-PAGEA7590,9010 ·
- Acrylamide 2K Standard grade, pure A1089,0500 · 79-06-1
- Acrylamide 4K - Solution (30 %) - Mix 29 : 1 A0951,1000 ·
Protein Biochemistry
Proteins are found in every cell. Their sequence is already mapped by the genetic code. Genes are read in the course of the so-called expression, so that the right proteins are available at the right time in the living being.
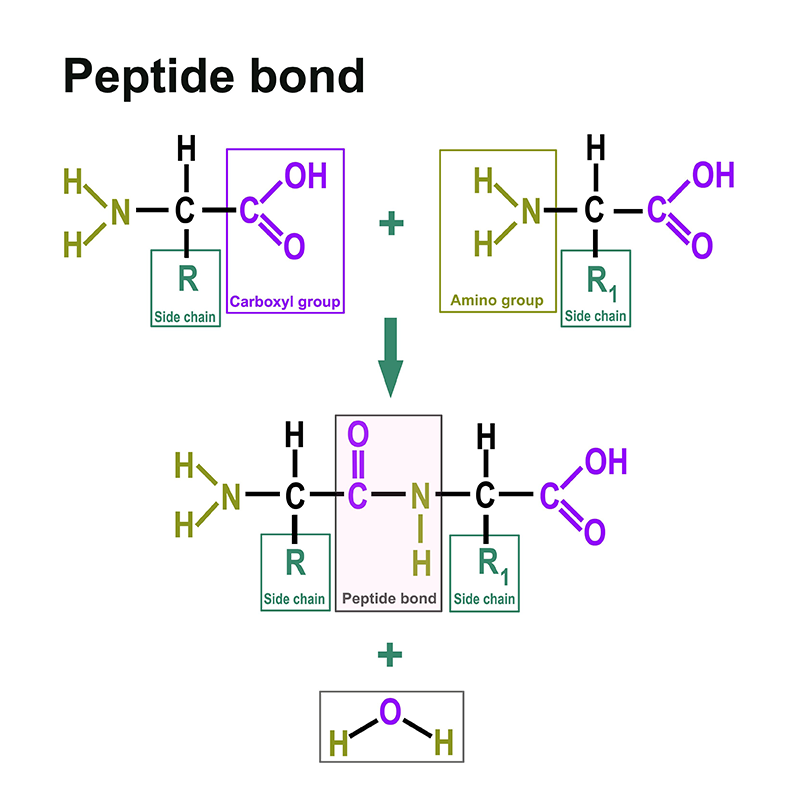
Proteins consist of amino acids. You can find a separate section on this here: https://www.itwreagents.com/rest-of-world/en/amino-acids. These amino acids are linked via so-called peptide bonds.
A peptide bond is a carboxylic acid-amide bond that connects two amino acids via the carboxy group of one amino acid and the amino group of the other amino acid. Most commonly, it is understood to be the bond between the respective functional groups in the α-position of two amino acids, i.e., between the C-1 of one amino acid and the N-2 of the other amino acid. The bond is formed by a condensation reaction with elimination of water.
The multiple combination possibilities of amino acids and also of peptides leads to an almost infinite possibility of structures and thus to many functions. Proteins can be enzymes and thus the catalysts of biology. For example, they can give structures to our cells, protect our genetic material and keep it packed. Proteins play a decisive role in our body's motor function and provide for the movement of the muscles in actin and myosin. The totality of the proteins of an organism is called proteome.
More precisely defined, the proteome comprises the totality of all proteins present in a cell or a living organism under defined conditions and at a defined time. In contrast to the rather static genome, the proteome and also the transcriptome are dynamic and can therefore change in their qualitative and quantitative protein composition due to changing conditions (environmental factors, temperature, gene expression, drug administration, etc.). The dynamics of the proteome can be visualised in the following example. A caterpillar and the butterfly that develops from it contain the same genome, but nevertheless differ externally due to a different proteome. The same is true for a tadpole and the frog that develops from it.
The changes in the proteome can sometimes occur very rapidly, for example through post-translational modifications such as the phosphorylation and dephosphorylation of proteins, which play a very important role in signal transduction.
Proteomics attempts to classify and catalogue all proteins in the organism and to decipher their functions. The blueprints of the proteins are found in the hereditary material. While the genetic material DNA only stores information, the protein molecules consisting of amino acids fulfil multiple tasks. They are the basic substance of life and, for example, as antibodies hey prevent diseases, and as enzymes they enable metabolism and provide movement in the form of skeleton, tendons and muscles.
The word proteome comes from the Australian researcher Marc Wilkins and was first mentioned on a slide in his presentation at the „2D Electrophoresis: from protein maps to genomes“ congress in Siena on 5 September 1994. The wording on the slide was: "Proteome: the PROTEin complement expressed by a genOME, cell or tissue”.
Similar to the Human Genome Organisation (HUGO), the researchers of the International Human Proteome Organisation (HUPO) share the workload worldwide. Germany is concentrating on research into brain proteins. In Germany, leading protein and proteomics scientists have also joined forces in the German Society for Proteome Research (DGPF) since 2001 in order to make optimal use of research capacities.
Subdivisions
Important subfields are the elucidation of protein-protein interactions, which mainly depend on the tertiary and quaternary structures of proteins and the interactions of their domains. Furthermore, protein purification and the quantitative analysis of protein expression also belong to the field of proteomics. It thus complements the data obtained in gene expression analysis and provides information on the components of metabolic pathways and molecular regulatory circuits. Protein engineering allows the modification of functions of recombinant proteins to adapt its properties.
Thus, the key techniques of proteomics support the elucidation of function and 3-D protein structure and the identification of individual proteins in mixtures.
Since all metabolic processes are carried out by proteins, therapeutic approaches such as new active agents against cancer, infections and certain nervous diseases are based on them. Diseases such as sickle cell anaemia, Alzheimer's disease, Huntington's chorea or Creutzfeldt-Jakob disease are based on incorrectly formed and clumping proteins. Therefore, if it is known which protein is responsible for a malfunction, it is possible to specifically develop a small molecule that docks onto this protein and prevents further malfunction. Recombinant proteins are used in industry in the form of detergent enzymes and biological pesticides. Biologists hope to gain better insights into the functioning of living organisms and life as such. Biophysicists expect a "molecular anatomy".
Systems Biology:
A new field of research that builds on proteomics is systems biology. This no longer attempts to look solely at the individual parts of a cell, for example, but tries to describe the interaction of all the individual parts within a system and its environment. In addition to proteomics, this requires mathematical models that simulate the system in silico (i.e. in computer models), but also those that allow this to be reproduced in experimental approaches.
Paleoproteomics
In addition to "old" DNA, fossil proteins can occasionally be isolated from fossil bones, which also allow conclusions to be drawn, for example, about their affiliation to a particular biological species. Paleoproteomics (from the Greek παλαιός palaiós, "old"), which is based on this, benefits in particular from the fact that some proteins are more stable than DNA over a longer period of time. In 2016, for example, the research group of Jean-Jacques Hublin at the Max Planck Institute for Evolutionary Anthropology used collagen samples dating back some 40,000 years to clarify that the archaeological culture of Châtelperronia is linked to Neanderthals and not to anatomically modern humans (Homo sapiens). In 2019, fossil proteins from dentin of the Xiahe mandible discovered in the highlands of Tibet in Baishiya Cave proved it to be of Denisova humans, and a few months later, 1.9 million-year-old dentin samples confirmed that the genus Gigantopithecus is an extinct "sister" taxon of orangutans. Back in 2015, collagen analyses revealed a closer relationship between the "South American ungulates" and the odd-toed ungulates, namely Macrauchenia and Toxodon, which were still present in the late Pleistocene. Previously, the exact relationships of the "South American ungulates" to other ungulate groups had been unclear and the subject of scientific debate. For the extinct rhinoceros Stephanorhinus, proteomes dating back some 200,000 to 400,000 and 1.8 million years, respectively, in 2017 and 2019 revealed a closer relationship to the woolly rhinoceros and thus to a closer circle of kinship around the modern Sumatran rhinoceros. The position could also be supported by genetic studies and had previously been assumed on anatomical grounds. Also in 2019, studies on proteins contributed to the systematic reclassification of fossil and recent sloths.
In 2015, for example, a study of 80-million-year-old bones of Brachylophosaurus canadensis, a member of the duck-billed dinosaur group, attracted worldwide media attention. The study revealed peptides that, due to their similarity to peptides found in chicken birds and ostriches living today, were interpreted as the remains of blood vessels.
Issues and trends
After some sobering experiences with genetic methods such as microarray analysis, some scientists are also somewhat skeptical about proteome research. Friedrich Lottspeich from the Max Planck Institute of Biochemistry in Martinsried, President of the German Society for Proteome Research (DGPF), warns against exaggerated hopes:
"For the human sector, the research is actually still too complex at the moment anyway [...] But for an analysis of yeast, which would be a good model system, again of course nobody wants to spend money."
The complexity arises from the many possibilities: According to Friedrich Lottspeich, humans are estimated to have several hundred thousand to millions of different proteins. A single gene produces an average of five to ten proteins, in some cases several hundred. To fully comprehend this complexity is a challenge that current methods are not yet up to. On the other hand, proteome research is developing rapidly. This is due in particular to a constant improvement in mass spectrometers, which are becoming ever more precise, sensitive and fast.
Another important step is the development of quantitative methods, such as the SILAC, iTRAQ, TMT or ICAT procedures based on the use of stable isotopes, or MeCAT metal coding, in which metals of different weights are used to label proteins and peptides from different protein samples. The latter allows for the first time in the multiplex approach the proteome-wide use of ultrasensitive elemental mass spectrometry (ICP-MS) (detection limit in the ppt to lower ppq range), which allows over 2 to 5 orders of magnitude higher sensitivity in protein quantification and has a linear dynamic measurement range of at least 6 to 8 orders of magnitude. MeCAT, in contrast to the other methods that 'only' quantify relatively at the peptide level, advantageously allows relative and even absolute quantification at the protein level, making protein species such as post-translationally modified proteins more amenable to quantification. Calibration of ICP-MS is performed using protein/peptide-independent metal standards. The need for protein-specific standard peptides is thus eliminated.
If quantitative proteome analysis is combined with other biological methods, it is also possible to make statements about the function of proteins (e.g. protein-protein interaction or post-translational modifications). Modern proteome research therefore now goes far beyond the mere cataloguing of proteins and attempts to understand complex mechanisms.
Simple applications
For researchers and developers, it is important to make proteins visible and to be able to quantify them. One of the most important techniques for this is polyacrylamide gel electrophoresis (PAGE). In PAGE, proteins are separated according to their size in an electric field. We offer everything you need to prepare the gels yourself. You will find a selection of acrylamide and bisacrylamide solutions, APS, TEMED and supporting materials DTT, β-mercaptoethanol, Coomassie and our popular protein marker.
Proteins are always predetermined in their mode of action and functional availability at sites of application in the organism that are predetermined by evolution. Therefore, the proteins must first be made available to the observer from the outside.
This happens on the one hand by physical methods, such as cell disruption by heat or chemically. For example, our product Tritidy G can help to isolate DNA, RNA and proteins from a sample simultaneously. However, detergents are particularly important in this context. Many well known detergents like SDS and Tween are available, but also more specific ones like Digitonin and Saponin, which are often used for membrane proteins. We would also like to point out our Triton X-100 replacement products, which offer greater protection for the user.
Triton X-100 Replacement Products:
| PRODUCT CODE | PRODUCT NAME | CAS NUMBER |
| A9778 | ECOSURF™ EH-9 (Triton X-100 replacement) |
64366-70-7 |
| A9779 | ECOSURF™ SA-9 (Triton X-100 replacement) |
|
| A9780 | TERGITOL™ 15-S-9 (Triton X-100 replacement) |
68131-40-8 |
The original, Triton X-100 has been classified as a high risk substance due to the REACH regulation and can no longer be sold easily.
The techniques of protein biochemistry are not only used to determine the natural states of proteins, but also to produce desired proteins by means of targeted manipulations. So-called expression systems are used for this purpose. One of the best known is the Lac-operon. Here, the genes for certain proteins are assembled by molecular biology using this operon. In bacteria (E. coli), the organism then produces the proteins. The whole process is induced by the substance isopropyl-β-D-thiogalactopyranoside (IPTG). We have a selection of IPTG available for our customers.
| PRODUCT CODE | PRODUCT NAME | CAS NUMBER |
| A1008 | IPTG BioChemica | 367-93-1 |
| A4773 | IPTG for molecular biology, dioxane free | 367-93-1 |
| A7211 | IPTG from plant origin galactose, dioxane free | 367-93-1 |
The biggest opponent of a protein is ... another protein. The degradation is catalyzed by proteases. These are found in all organisms and therefore often also in the samples of users/scientists. Since it would often be uneconomical to achieve a direct separation, so-called inhibitors are used. These interfere with the catalytic activity of the proteases so that they cannot become active and other enzymes remain intact.
Sometimes, however, it is precisely the protein degradation that is desired. For example, if one only wants to examine the nucleic acids in a sample. The proteins can then be interfering. A well-known example might be certain test kits for COVID-19. Proteinase K is a protease that catalyzes the degradation of proteins. We offer high quality Proteinase K in lyophilized and already dissolved ready-to-use form.
Other important products that are used are albumins and co-factors such as NAD and NADPH. They may be needed as stabilizers or as coenzymes in various reactions.
The field of proteomics will remain exciting for a very long time. It shows us the true actual state of each cell. Many assays will still lead to some groundbreaking discoveries here.
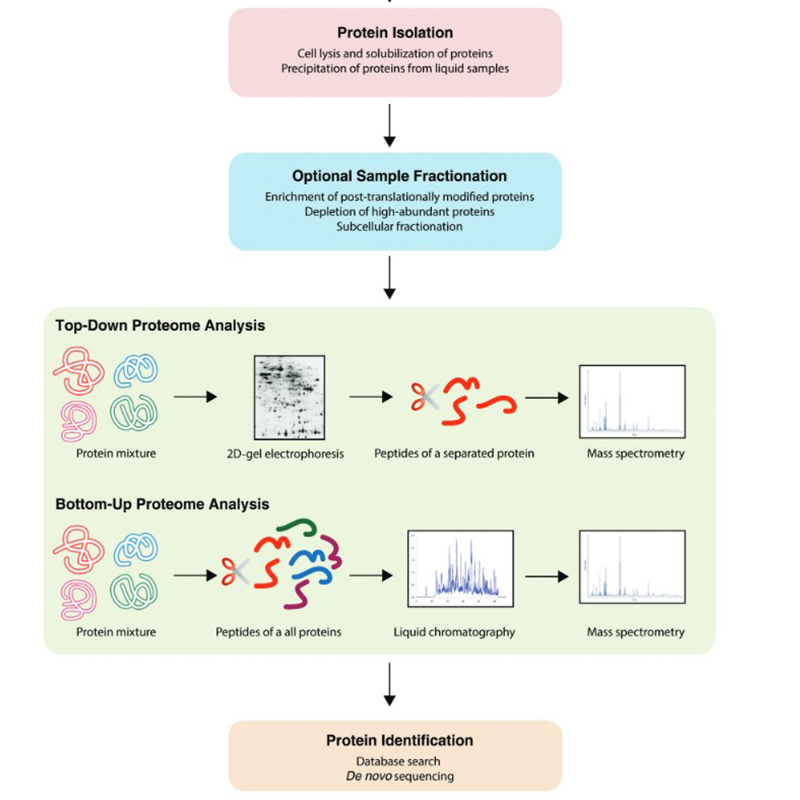
In addition to the techniques already mentioned above, a variety of workflows can be found in the field of proteomics. These can take place botton-up or top-down:
.png)
Proteomics is also carried out by means of models in which cell or animal-based models are used. 2D gel electrophoresis and labelling play a role. Large steps in the field of proteomics were made by chromatographic methods coupled with mass spectrometry. For this purpose we offer the most common solvents.
Methods commonly used in proteomics today include chromatography, mass spectrometry, MALDI imaging, one-dimensional polyacrylamide electrophoresis (1D PAGE), two-dimensional polyacrylamide gel electrophoresis (2D PAGE), microarrays, amino acid analysis (AAA), cell culture and, most importantly, a wide use of bioinformatic models.
Particularly in tumour diagnostics, qualitative and quantitative proteomics is used and allows conclusions to be drawn for prognoses and therapies for individual patients.
Proteins are also of great importance in immunology. Our antibodies are also special forms of proteins.
The complexity of proteomics arises from the very many possibilities: According to Friedrich Lottspeich (Austrian biochemist), humans are estimated to have several hundred thousand to millions of different proteins. A single gene produces on average five to ten proteins, in some cases several hundred. To fully grasp this complexity is a challenge that current methods are not yet up to. On the other hand, proteome research is developing rapidly. This is due in particular to a constant improvement in mass spectrometers, which are becoming ever more precise, sensitive and fast.
An overview of important articles, suitable for the research of protein biochemistry (proteomics) can be found below:
| PRODUCT CODE | PRODUCT NAME | CAS NUMBER |
| A1088 | ABTS® BioChemica | 30931-67-0 |
| A4983 | Acrylamide solution (30%) - Mix 29 : 1 for molecular biology | |
| A3626 | Acrylamide solution (30%) - Mix 37.5 : 1 for molecular biology | |
| A3658 | Acrylamide solution (40%) - Mix 19 : 1 for molecular biology | |
| A0385 | Acrylamide solution (40%) - Mix 29 : 1 for molecular biology | |
| A4989 | Acrylamide solution (40%) - Mix 37.5 : 1 for molecular biology | |
| A7582 | Acrylamide 2K solution (18%) for denaturing DNA-PAGE | |
| A7590 | Acrylamide 2K solution (8%) for denaturing DNA-PAGE | |
| A1089 | Acrylamide 2K Standard grade, extrapure | 79-06-1 |
| A0951 | Acrylamide 4K solution (30%) - Mix 29 : 1 | |
| A1672 | Acrylamide 4K solution (30%) - Mix 37.5:1 | |
| A1577 | Acrylamide 4K solution (40%) - Mix 37.5:1 | |
| A1090 | Acrylamide 4K ultrapure | 79-06-1 |
| A1421 | AEBSF Hydrochloride BioChemica | 30827-99-7 |
| A0850 | Albumin (BSA) EIA and RIA grade | 9048-46-8 |
| A2244 | Albumin (BSA) Fraction V (pH 5.2) | 9048-46-8 |
| A1391 | Albumin (BSA) Fraction V (pH 7.0) | 9048-46-8 |
| A6588 | Albumin (BSA) Fraction V (pH 7.0) for Western blotting | 9048-46-8 |
| A4344 | Albumin crude from chicken egg white | 9006-59-1 |
| A1523 | 4-Aminoantipyrine BioChemica | 83-07-8 |
| A7708 | AppliCoat Plate Stabilizer | |
| A2132 | Aprotinin BioChemica | 9087-70-1 |
| A2568 | Avidin BioChemica | 1405-69-2 |
| A1117 | BCIP BioChemica | 6578-06-9 |
| A3636 | Bisacrylamide for molecular biology | 110-26-9 |
| A3417 | CheLuminate-HRP PicoDetect | |
| A2144 | Chymostatin | 9076-44-2 |
| A1101 | DTT BioChemica | 3483-12-3 |
| A2948 | DTT for molecular biology | 3483-12-3 |
| A3668 | DTT solution 1 mol/L (1 M) for molecular biology | 3483-12-3 |
| A1007 | X-Gal BioChemica | 7240-90-6 |
| A4978 | X-Gal for molecular biology | 7240-90-6 |
| A2243 | L-Glutathione oxidized BioChemica | 27025-41-8 |
| A9782 | L-Glutathione reduced (USP) pure, pharma grade | 70-18-8 |
| A2084 | L-Glutathione reduced BioChemica | 70-18-8 |
| A1008 | IPTG BioChemica | 367-93-1 |
| A4773 | IPTG for molecular biology, dioxane free | 367-93-1 |
| A7211 | IPTG from plant origin galactose, dioxane free | 367-93-1 |
| A2185 | Luminol | 521-31-3 |
| A1108 | β-Mercaptoethanol for molecular biology | 60-24-2 |
| A1243 | NBT BioChemica | 298-83-9 |
| A1272 | 2-Nitrophenyl-β-D-Galactopyranoside BioChemica | 369-07-3 |
| A1028 | 4-Nitrophenyl-β-D-Glucuronide BioChemica | 10344-94-2 |
| A0830 | Nonfat dried milk powder | |
| A2205 | Pepstatin A | 26305-03-3 |
| A0999 | PMSF BioChemica | 329-98-6 |
| A2935 | Ponceau S - Solution | |
| A8889 | Protein Marker VI (10 - 245) prestained | |
| A1495 | Streptavidin ultrapure | 9013-20-1 |
| A1148 | TEMED | 110-18-9 |
| A3840 | 3,3',5,5'-Tetramethylbenzidine BioChemica | 54827-17-7 |
| A1828 | Trypsin inhibitor from soybean > 7000 BAEE | 9035-81-8 |